-Search query
-Search result
Showing 1 - 50 of 1,054 items for (author: ho & ky)

EMDB-43751: 
TRPM7 structure in complex with anticancer agent CCT128930 in closed state
Method: single particle / : Nadezhdin KD, Sobolevsky AI

PDB-8w2l: 
TRPM7 structure in complex with anticancer agent CCT128930 in closed state
Method: single particle / : Nadezhdin KD, Sobolevsky AI

EMDB-29907: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v); consensus map with only Fab 1G01 resolved
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

EMDB-29908: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), locally refined map
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

EMDB-29909: 
Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2)
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

PDB-8gat: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), based on consensus cryo-EM map with only Fab 1G01 resolved
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

PDB-8gau: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v)
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

PDB-8gav: 
Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2)
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M
EMDB-35377: 
Cryo-EM structure of GPR156 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35378: 
Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35380: 
Cryo-EM structure of GPR156-miniGo-scFv16 complex
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35382: 
Cryo-EM structure of GPR156A/B of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35389: 
Cryo-EM structure of GPR156C/D of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35390: 
Cryo-EM structure of G-protein free GPR156
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ieb: 
Cryo-EM structure of GPR156 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iec: 
Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ied: 
Cryo-EM structure of GPR156-miniGo-scFv16 complex
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iei: 
Cryo-EM structure of GPR156A/B of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iep: 
Cryo-EM structure of GPR156C/D of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ieq: 
Cryo-EM structure of G-protein free GPR156
Method: single particle / : Shin J, Park J, Cho Y

EMDB-40470: 
Pendrin in complex with chloride
Method: single particle / : Wang L, Hoang A, Zhou M

EMDB-40479: 
Pendrin in complex with iodide
Method: single particle / : Wang L, Hoang A, Zhou M

EMDB-40483: 
Pendrin in complex with Niflumic acid
Method: single particle / : Wang L, Hoang A, Zhou M

EMDB-40507: 
Pendrin in complex with bicarbonate
Method: single particle / : Wang L, Hoang A, Zhou M

EMDB-42588: 
Pendrin in apo
Method: single particle / : Wang L, Hoang A, Zhou M

EMDB-37631: 
Hepatitis B virus capsid (HBV core protein)
Method: single particle / : Yip RPH, Lai LTF, Lau WCY, Ngo JCK, Kwok DCY

EMDB-37634: 
SR protein kinase 2 bound at 2-fold vertex of Hepatitis B virus capsid
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK

EMDB-38062: 
Hepatitis B virus capsid in complex with SR protein kinase 2
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-15214: 
Endogenous yeast L-A helper virus identified from native cell extracts
Method: single particle / : Schmidt L, Kyrilis F, Hamdi F, Semchonok DA, Kastritis PL

EMDB-41048: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

PDB-8t5c: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

EMDB-34022: 
Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide
Method: single particle / : Naganuma M, Ehara H, Kim D, Nakagawa R, Cong A, Bu H, Jeong J, Jang J, Schellenberg MJ, Bunch H, Sekine S

PDB-7yq8: 
Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide
Method: single particle / : Naganuma M, Ehara H, Kim D, Nakagawa R, Cong A, Bu H, Jeong J, Jang J, Schellenberg MJ, Bunch H, Sekine S
EMDB-34174: 
Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human Atypical chemokine receptor 2, ACKR2 (D6R)
Method: single particle / : Maharana J, Sarma P, Yadav MK, Banerjee R, Shukla AK
EMDB-36078: 
Structure of beta-arrestin2 in complex with M2Rpp
Method: single particle / : Maharana J, Sano FK, Shihoya W, Banerjee R, Nureki O, Shukla AK
EMDB-36081: 
Structure of beta-arrestin2 in complex with D6Rpp (Local Refine)
Method: single particle / : Maharana J, Sarma P, Yadav MK, Chami M, Banerjee R, Shukla AK
EMDB-36082: 
Structure of beta-arrestin1 in complex with D6Rpp
Method: single particle / : Maharana J, Sarma P, Yadav MK, Chami M, Banerjee R, Shukla AK
EMDB-36090: 
Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local refine, cross-linked)
Method: single particle / : Maharana J, Sano FK, Shihoya W, Banerjee R, Nureki O, Shukla AK
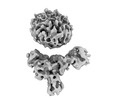
EMDB-36091: 
Muscarinic receptor (M2R) in complex with beta-arrestin1 (Low resolution full map, Cross-linked)
Method: single particle / : Maharana J, Sano FK, Shihoya W, Banerjee R, Nureki O, Shukla AK
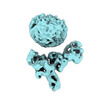
EMDB-36093: 
Muscarinic receptor (M2R) in complex with beta-arrestin1 (Low resolution full map, non-crosslinked)
Method: single particle / : Maharana J, Sano FK, Shihoya W, Banerjee R, Nureki O, Shukla AK
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search








 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
